CryoSPARC
"CryoSPARC is a state of the art scientific software platform for cryo-electron microscopy (cryo-EM) used in research and drug discovery pipelines."
M3 offers a "General Access (GA)" CryoSPARC that is available to any M3 user upon request.
Important note on data security
Since the backend GA CryoSPARC user has access to all CryoSPARC projects, all users can read each others' files. If you have sensitive data, our GA CryoSPARC is not suitable for you.
Requesting access
- Submit this form to request access to M3's GA CryoSPARC.
- Wait up to 2 weeks for your request to be processed.
- You should receive an email confirming your access. This email will contain instructions on how to log into CryoSPARC for the first time and telling you to reset your password. Note your CryoSPARC credentials are different from your HPC ID or usual university credentials.
Usage
- Start a lightweight Strudel desktop
- In the Strudel desktop, open a browser, e.g. FireFox.
- Type
http://m3t101:39000into the browser's URL bar, and pressEnter. - Sign in with your CryoSPARC-specific username and password.
- Use CryoSPARC!
For the desktop, you only need to request minimal resources to run the browser. E.g.
cpus-per-task=2--mem=8G- No GPUs!
Requesting significant resources for the deskopt is pointless because CryoSPARC won't actually be using them. Instead, CryoSPARC submits its own job requests on behalf of you from the web interface.
Which directories can CryoSPARC use?
The filepaths will look slightly different when you're using CryoSPARC vs when you're not using
CryoSPARC. Replace <PROJECT_ID> below with your M3 project ID.
| Inside CryoSPARC? | Filepaths |
|---|---|
| Yes | /cryosparc/<PROJECT_ID> |
| No | /fs04/scratch2/<PROJECT_ID>/cryosparc |
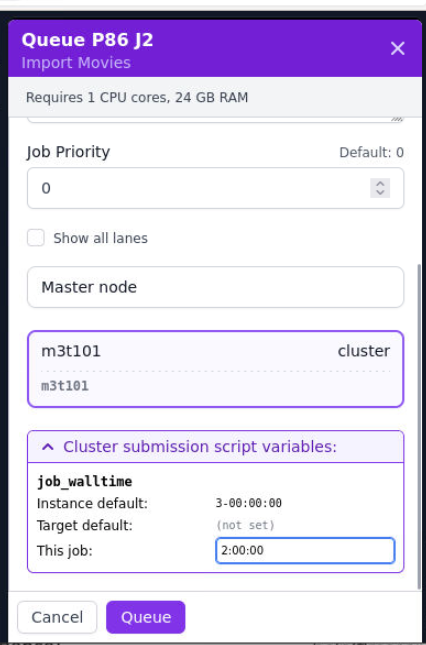
Setting a job's time limit
This is generally not necessary, but you can manually adjust the time limit for your jobs in our CryoSPARC:
- Set up your CryoSPARC job as usual, up to the queuing step.
- Click on "Cluster submission script variables".
- Modify the
job_walltimefield for "This job". This value should be of any format accepted by Slurm'ssbatchcommand for the--timefield.
See the screenshot below for an example.
Troubleshooting
sbatch: error: Required nodes outside of the reservation
If a maintenance period is approaching, you may see the following error message in CryoSPARC:
sbatch: error: Required nodes outside of the reservation
sbatch: error: Batch job submission failed: Requested node configuration is not available
This occurs if your job's runtime would overlap with the maintenance period. A workaround is to reduce your CryoSPARC job's time limit such that it can complete before the maintenance period begins. Follow the instructions in Setting a job's time limit to reduce your time limit.
As the maintenance period draws closer, this workaround will eventually stop working. At this point, you will simply have to wait until the maintenance is completed before you can submit any new jobs to CryoSPARC. Note that even if, for example, the maintenance begins in 8 hours and you set the time limit to only 1 hour, you may still see the above error message since other jobs (including those from other users) could still be in the job queue.
Particle picking issues (Topaz)
You may encounter errors when using Topaz in CryoSPARC or RELION. A wrapper script was created to work around this.
To use this script in CryoSPARC, set the Path to Topaz executable to
/usr/local/topaz/0.2.5-20240320/topaz-relion
You should be able to find a similar input to specify the Topaz executable in RELION.
Topaz may also be an alternative for CryoSPARC's DeepPicker which (as of writing this) has a pretty major bug at inference time.